Low-resolution membrane protein structure from small-angle scattering data.
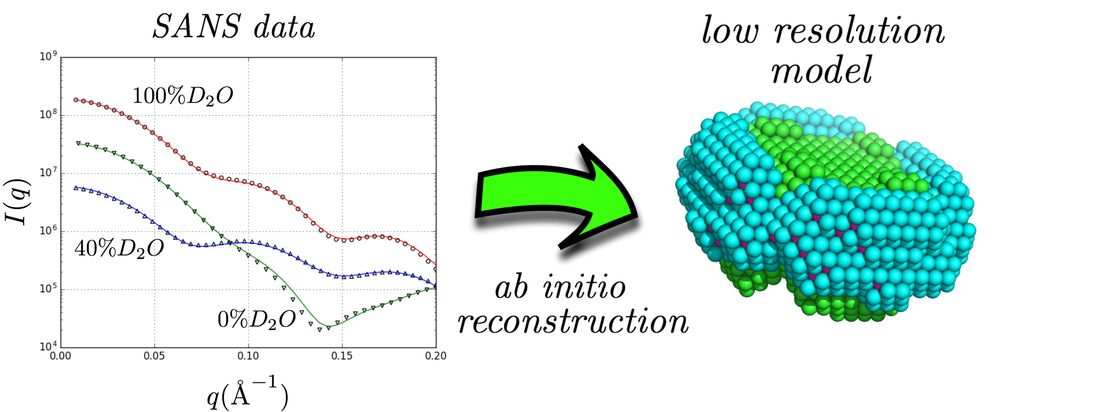
Attacking the problem of membrane protein structure has proved notoriously difficult, due to the limited applicability of established experimental techniques for this class of systems. This was also the case for small-angle scattering of x-rays and neutrons. Popular analysis tools (like ab-initio methods) cannot be applied in a straightforward manner due to the need of detergent molecules or other molecular assemblies for the solubilisation of membrane proteins, that lead to extra contributions to the scattering signal. In a new paper published in the Biophysical Journal we illustrate that Small-Angle Neutron Scattering data at different solvent contrasts (i.e. D2O/H2O ratios) permit through the use of an appropriate model and minimization procedure the reconstruction of the low resolution structure of the different parts of detergent / protein complexes. This developed model attempts to fit the experimental data using a multiphase coarse-grained approach that encompasses a set of physical constraints adapted to the anticipated assembly of detergents around the protein surface. A related software program (DANVILLE) was developed and is provided freely to the academic community.